| Home | Up | Questions? |
There are several organizations that will do DNA analysis for people.
This costs $199 for both Ancestry and Health analysis. I
signed up in March 2017 and sent them a tube of spit on 25
March. I had e-mailed results
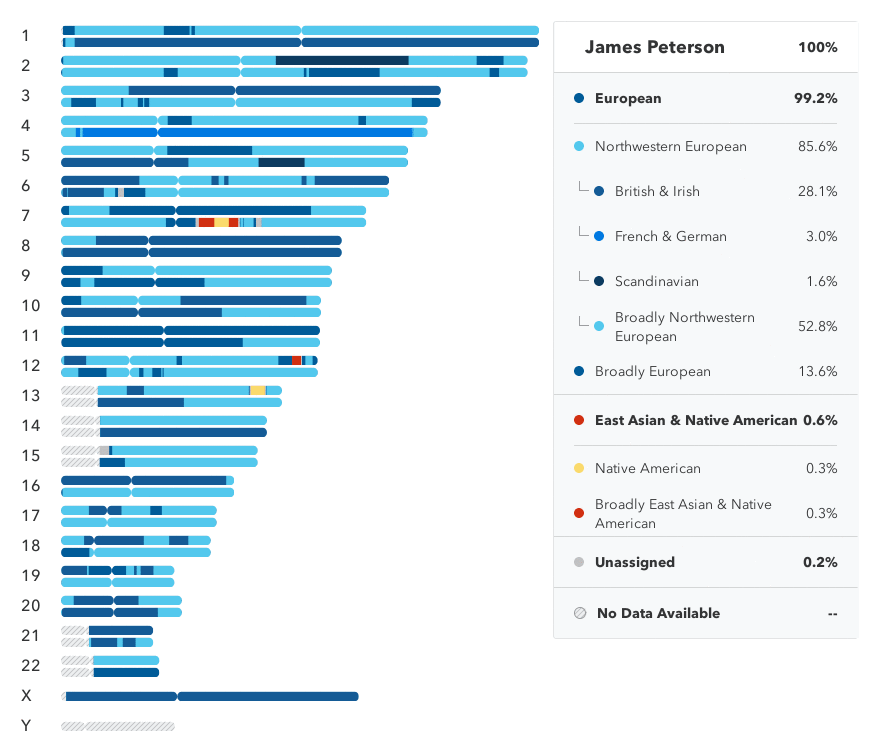
on 16 April. The results show that I am basically a very
uninteresting set of DNA, almost completely European (99.2%).

This is a study run by the University of Michigan. The analysis did not cost money, but they required answers to lots of questions that may help them find correlations between DNA and health/life properties. I signed up on 4 April 2017, sent them a spit kit that they got on 2 May, and had results on 11 August.
Again, the results show I am an uninteresting set of DNA. I am almost solely
European (93%) with a tad of Central and South Asia (6%) and Native America (1%).
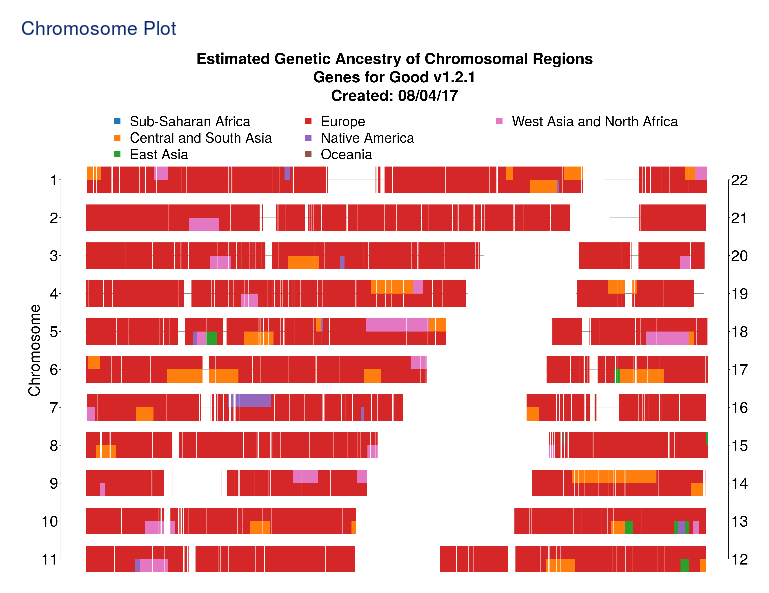
Each of the two organizations will allow me to down load the raw DNA files. 23andme has their own format for the data; Genes for Good downloads several files, one of which is in the 23andme format (more or less). These files show what they have actually sequenced. There are 23 chromosomes pairs. Each of the pairs is a strand or chain of amino acids. Actually, of course, each chromosome is a (twisted) double strand. There are only 4 amino acids used:
So we have 23 chromosomes. Each chromosome is a chain of amino acids. There are 4 amino acids. The chromosomes are really long. For example, chromosome 1 has at least 249,218,992 amino acids. The complete genome for people (all 23 pairs) has at least 7 billion amino acids. Of course, most of these are the same for all people. Eye color may vary, for example, but we all have eyes and they are all built pretty much the same, so there are a lot of amino acids that define the eye (and everyone has the same ones for that) and a few that define the color (and for those people vary, although not a lot).
So there is a "reference genome". The current one in use is called GRCh37. And all that the DNA files have is where my genome varies from the reference genome. And then only the places that they sequence, probably not all the places it could vary from GRCh37. As a result the files are relatively small -- they don't have the complete 7 billion amino acids. For 23andme, I have 610,362 items; for GenesForGood, I have 563,243 items.
Each of these items is a line in a file with 2 basic pieces of information: where the item is located -- which chromosome and what position on that chromosome -- and what the amino acid is at that location (A,C,G,T). And of course you have two pairs -- one from your Mother and one from your Father -- so you get two amino acids at that location.
There are exceptions. The 23rd chromosome is the sex chromosome, and is either a pair of two X chromosomes (female) or one X and one Y (male). You get one from your Mother (always an X) and one from your Father (either an X if you are female or a Y if you are male). These get listed separately. The Y chromosome, for example, is very short. Since these are listed separately, you only have one amino acid listed at each location for the X and Y.
One other exception. Most of your DNA is in the 23 pairs of chromosomes. But there is another part of the cell called the mitochondria; it has it's own DNA (mitochondrial DNA). The mitochondria from a cell is duplicated whenever the cell splits; the original copy is in the egg. So your mitochodrial DNA comes directly from your Mother. It's fairly short, but is distinct from the basic 23 chromosomes.
So I have information for 22 paired chromosomes, plus my X, Y, and MT (mitochondrial) DNA. From each of the two organizations. Are they the same?
The two organizations did not sequence the same locations. If we look at just the chromosome and position information, they both sequenced only 147,182 common locations (out of 610,362 and 563,243), so not a lot of overlap (only about 25%), which is somewhat surprising to me.
Of the 147,182 common locations sequenced, all but 28 are the
same. Of these 28, we have confusion over which of two amino
acids it should be as:
| Number of times | possibly | or maybe |
|---|---|---|
| 2 | A | C |
| 11 | A | G |
| 2 | C | G |
| 12 | C | T |
| 1 | G | T |
It would seem to me that there are two possible explanations for these differences:
Uploaded my 23andme DNA data to GEDmatch. They say: Assigned kit number: M724923 Write this number down. File identified as 23andMe kit type V4
uploaded the GenesForGood DNA data to GEDmatch. There seems to be a difference of opinion on the "name" part at the begining of a line. Each 23andme format record is a "name" "number" "location" "genotype". The name is also called an RSID, a unique ID. But GFG makes some of them up with a syntax of number:offset_genotype1/genotype2 to say that on chromosome "number", at the offset, we normally expect to see genotype1 or genotype2. Sometimes genotype2 is "-", or either of them could be a string of ATCG characters.
GEDmatch does not like / in the name. And expects the file to be sorted by chromosome number (I suspect it wants it sorted by "number offset".) So it took some futzing the file to get it to upload correctly. It was assigned kit number M663610.
Not unsurprisingly, GEDmatch thinks the two DNA kits are all but identical.
| Home | Comments? |